To view your price please login or contact us
qPCRBIO Probe Mix
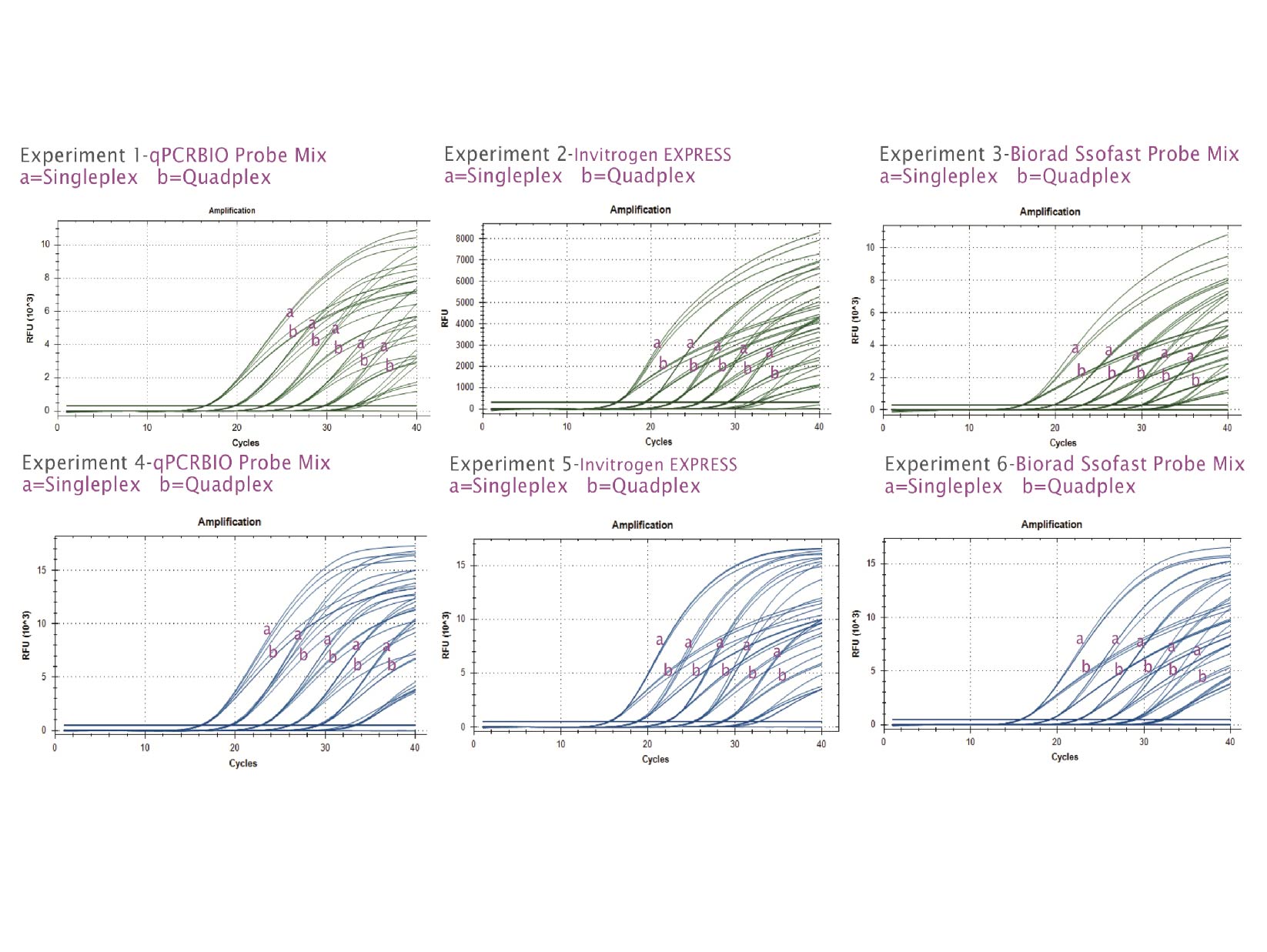
Our universal probe qPCR kit is designed to give superior sensitivity and specificity in all probe-based real-time PCR assays, including TaqMan®, Scorpions® and molecular beacon probes.
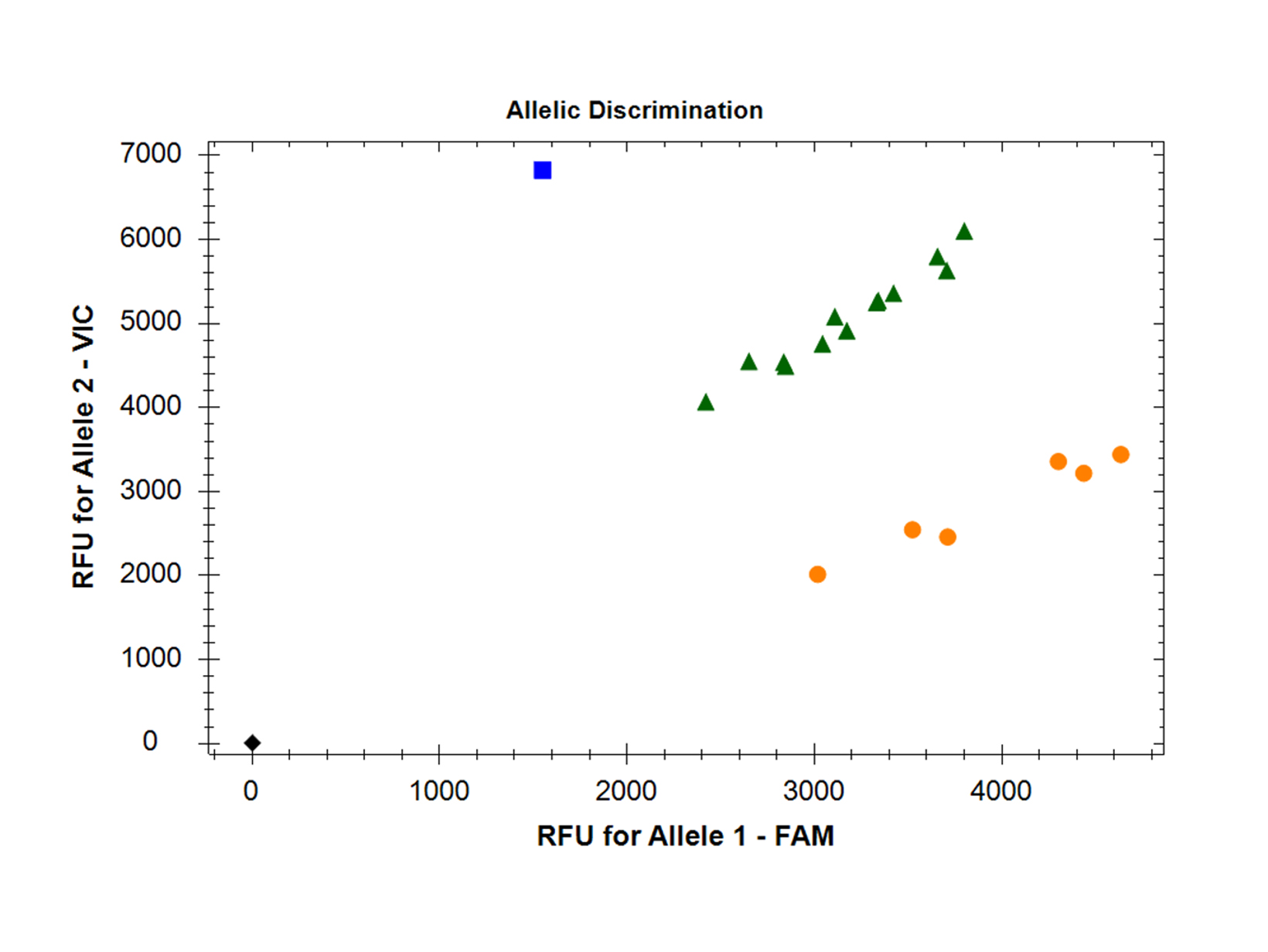
Reliably perform singleplex or multiplex expression studies and diagnostic assays with this robust, market-leading probe kit. The optimised mastermix is also suitable for qPCR-based genotyping and allelic discrimination studies. This mix is also available with a blue dye for easy visualisation as qPCRBIO Probe Blue Mix.
Features
- High efficiency in multiplex reactions
- Rapid extension rate for early Ct values
- Market leading sensitivity – increased limit of detection
- Efficient amplification from GC and AT-rich sequences
- Antibody-mediated hot start technology
- Compatible with all real-time PCR platforms
- Also available as an easy-to-see blue mix
More Information
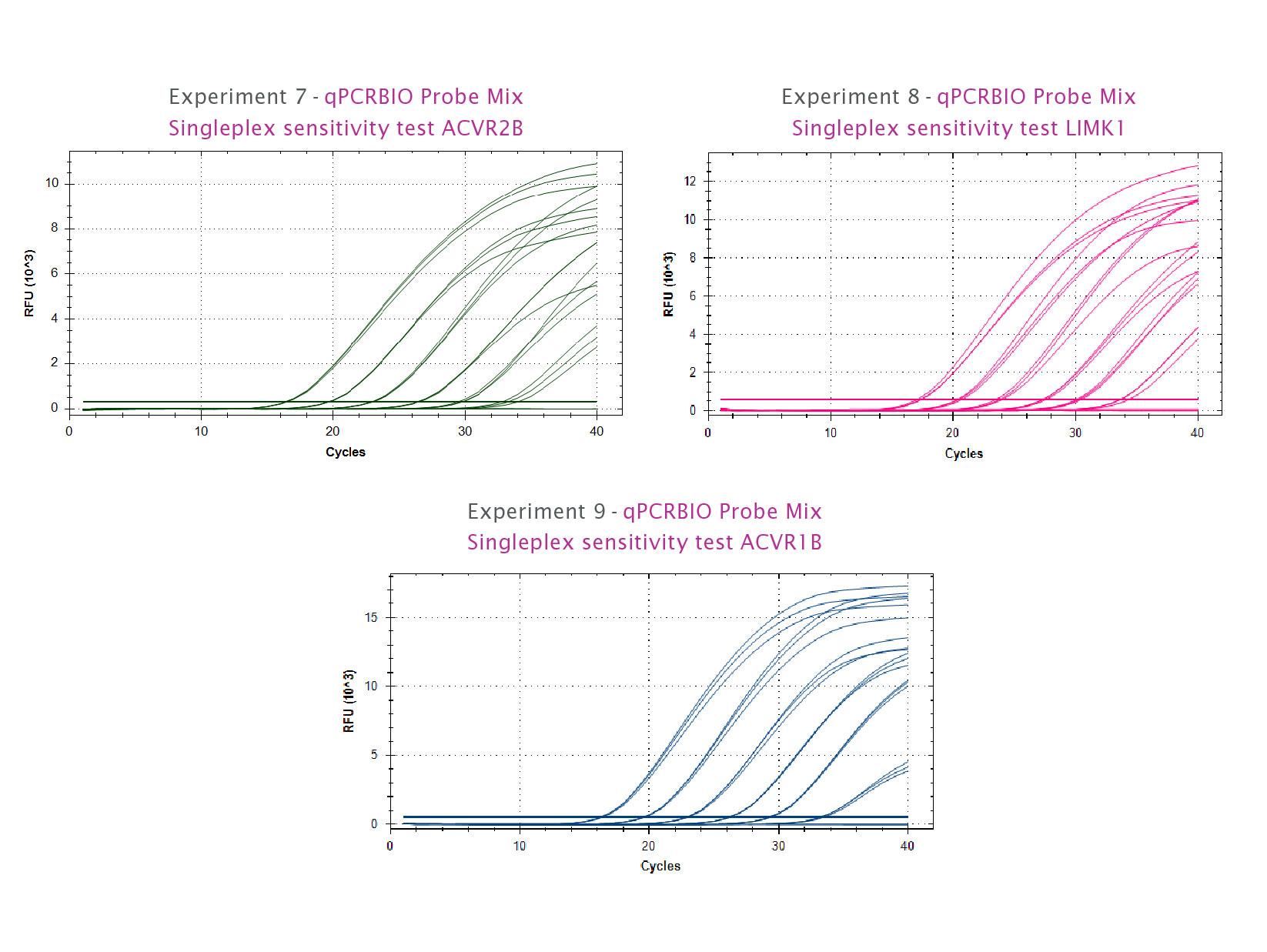
qPCRBIO Probe Mix is the robust choice for all your probe-based real-time PCR needs. Our universal kit can be used to reliably detect extremely low copy number targets and quantify any DNA template including genomic, cDNA and viral sequences.
By combining antibody-mediated hot start technology with the latest advances in buffer chemistry, we offer market-leading performance with minimal or no optimisation required. The enhanced sensitivity makes qPCRBIO Probe Mix the perfect choice for multiplexing.
Use our qPCR Selection Tool to find out which ROX variant is compatible with your instrument.
Applications
- Absolute quantification
- Relative gene expression analysis
- TaqMan®, Scorpions® and molecular beacon probes
- Detection of extremely low copy number targets
- Diagnostic real-time PCR
- Genotyping & allelic discrimination
Specifications
qPCRBIO Probe Mix Lo-ROX
Component
100 Reactions
500 Reactions
2000 Reactions
5000 Reactions
5000 Reactions
2x qPCRBIO Probe Mix Lo-ROX
1 x 1mL
5 x 1mL
20 x 1mL
1 x 50mL Bottle
50 x 1mL in Pouch
qPCRBIO Probe Mix Hi-ROX
Component
100 Reactions
500 Reactions
2000 Reactions
5000 Reactions
5000 Reactions
2x qPCRBIO Probe Mix Hi-ROX
1 x 1mL
5 x 1mL
20 x 1mL
1 x 50mL Bottle
50 x 1mL in Pouch
qPCRBIO Probe Mix No-ROX
Component
100 Reactions
500 Reactions
2000 Reactions
5000 Reactions
5000 Reactions
2x qPCRBIO Probe Mix No-ROX
1 x 1mL
5 x 1mL
20 x 1mL
1 x 50mL Bottle
50 x 1mL in Pouch
qPCRBIO Probe Mix Separate-ROX
Component
100 Reactions
500 Reactions
2000 Reactions
5000 Reactions
5000 Reactions
2x qPCRBIO Probe Mix No-ROX
1 x 1mL
5 x 1mL
20 x 1mL
1 x 50mL Bottle
50 x 1mL in Pouch
50μM ROX Additive
1 x 200μL
1 x 200μL
4 x 200μL
2 x 520μL
2 x 520μL
qPCRBIO Probe Mix Lo-ROX
Component
2x qPCRBIO Probe Mix Lo-ROX
100 Reactions
1 x 1mL
500 Reactions
5 x 1mL
2000 Reactions
20 x 1mL
5000 Reactions
1 x 50mL Bottle
5000 Reactions
50 x 1mL in Pouch
qPCRBIO Probe Mix Hi-ROX
Component
2x qPCRBIO Probe Mix Hi-ROX
100 Reactions
1 x 1mL
500 Reactions
5 x 1mL
2000 Reactions
20 x 1mL
5000 Reactions
1 x 50mL Bottle
5000 Reactions
50 x 1mL in Pouch
qPCRBIO Probe Mix No-ROX
Component
2x qPCRBIO Probe Mix No-ROX
100 Reactions
1 x 1mL
500 Reactions
5 x 1mL
2000 Reactions
20 x 1mL
5000 Reactions
1 x 50mL Bottle
5000 Reactions
50 x 1mL in Pouch
qPCRBIO Probe Mix Separate-ROX
Component
2x qPCRBIO Probe Mix No-ROX
50μM ROX Additive
100 Reactions
1 x 1mL
1 x 200μL
500 Reactions
5 x 1mL
1 x 200μL
2000 Reactions
20 x 1mL
4 x 200μL
5000 Reactions
1 x 50mL Bottle
2 x 520μL
5000 Reactions
50 x 1mL in Pouch
2 x 520μL
Reaction Volume
Storage
20μL
On arrival, products should be stored between -30 and -15°C. If stored correctly the kit will retain full activity for 12 months.
Reaction Volume
20μL
Storage
On arrival, products should be stored between -30 and -15°C. If stored correctly the kit will retain full activity for 12 months.
Instrument Compatibility
This product is compatible with all standard and fast cycling qPCR instruments. Use our qPCR Selection Tool to find out which ROX variant is compatible with your instrument.
Documents
Product Flyers
Product Manuals
Material Safety Data Sheets
Certificate of Analysis Finder
FAQs
Can products generated with qPCRBIO Probe Mixes be digested, cloned, and sequenced?
Yes, PCR products generated with qPCRBIO Probe Mixes have the same characteristics as PCR products generated with wild-type Taq polymerase. They may be sequenced or digested with restriction endonucleases using standard protocols. Products are 3′-d(A)-tailed and may be used for TA cloning or may be blunt-ended or digested with restriction enzymes prior to cloning. For best results, we recommend purifying the PCR products using any standard PCR clean-up kit.
Can ROX have a negative impact on the reaction?
ROX (6-carboxy-X-rhodamine) is used as a passive reference dye in ROX-dependent real-time PCR instruments to normalize for variations of fluorescence levels that can arise mainly due to optical path variations among wells. Normalisation of the fluorescence intensity (Rn) is done in real-time PCR software by dividing the emission intensity of the specific signal by the emission intensity of ROX.
ROX does not take part in the PCR reaction and its fluorescence levels are not proportional to the quantity of DNA in each well, so the addition of this fluorophore to a mix provides a constant fluorescent signal during amplification.
Different types of real-time PCR instruments requiring a passive reference standard have different optimal concentrations of ROX, mainly due to the different optical configurations of each system (i.e. the different type of excitation source and optics used).
The addition of either too little or too much ROX would result in a very noisy signal impacting on the results of the reaction. Therefore, it is extremely important for the user to:
- Determine the correct ROX concentration to optimise real-time PCR results, and
- Check the ROX settings on the software used to set up the reaction
A useful selection tool for the most commonly used systems can be found here.
Can the activation step at 95°C be increased to 15 minutes if DNA needs to be isolated from bacteria (colony PCR) or from viral particles?
Yes. The usual activation time of 2 minutes is not a long enough time to ensure release of the DNA from the bacteria or virus sample. The activation step can be increased to 15 minutes without affecting the reaction yield.
Do qPCRBIO Probe Mixes contain the FAM dye in it?
No. Apart from ROX, there is no other dye in our mixes. You can therefore use any fluorophore-conjugated probe for your reaction. Keep in mind that if you’re using qPCRBIO Probe Blue Mixes, refer to Table 1 in the respective product manuals to ensure the blue dye will not interfere with the fluorescence of your fluorophore.
Is it normal if the fluorescence of qPCRBIO Probe Mixes differs from the one obtained with competitors’ products?
Different products could give a different plateau of fluorescence. However, this has no impact on quantification accuracy and Ct values will not differ among products.
Is the storage of sample DNA in 1x TE (10 mM Tris-HCl / 1 mM EDTA) buffer compatible with subsequent qPCR using qPCRBIO Probe Mixes?
Yes, this storage buffer is compatible. The EDTA will chelate some of the magnesium in the mix, but not significantly enough to affect the reaction.
What are the ROX concentrations in qPCRBIO Probe Mixes?
The qPCRBIO Probe Mixes that contain passive reference dyes come in different formulations, each with a different concentration of the passive reference dye:
- qPCRBIO Probe Mix Lo-ROX (PB20.21) contains 112 nM ROX.
- qPCRBIO Probe Mix Hi-ROX (PB20.22) contains 1.12 µM ROX.
- qPCRBIO Probe Mix No-ROX (PB20.23) does not contain ROX.
- qPCRBIO Probe Mix Separate-ROX (PB20.24) 2x mix contains no ROX and include a separate tube of 50 µM ROX additive. This enables you to choose what concentration of ROX you’d like to use.
You can use our qPCR Selection Tool under the Resources drop-down menu to determine which of our mixes are best suited for your qPCR machine.
What is included in qPCRBIO Probe Mixes?
qPCRBIO Probe Mixes are ready to use qPCR 2x Mastermixes. You only need to add primers, template DNA and PCR grade water during reaction set up.
What is ROX?
ROX is a passive reference dye which means it does not take part in the PCR reaction. It is used to normalise non-PCR related fluctuations in fluorescence.
What is the concentration of dNTPs in qPCRBIO Probe Mixes?
All qPCRBIO Probe Mixes contain dNTPs at a concentration of 2 mM (0.5 mM each). This means the final concentration in the reaction is 1 mM (0.25 mM each).
What is the MgCl2 concentration in qPCRBIO Probe Mixes?
All qPCRBIO Probe Mixes contain MgCl2 at a concentration of 12 mM. This means the final concentration in the reaction is 6 mM.
What should be considered if the normalisation of signal with ROX appears to be lower relative to a competitor’s mix?
Ensure you are using the right concentration of ROX because different instruments require different ROX concentrations. For example, if a qPCRBIO Probe Mix Hi-ROX is used with an instrument requiring a Lo-ROX mix, the software will normalise the qPCRBIO signal against the Hi-ROX level. This will significantly reduce the fluorescence level of the qPCRBIO mix relative to competitors’ mixes, if in that case the Lo-ROX mix was chosen for the competitor.
When comparing mixes from different manufacturers, it is better to carry out separate runs or turn the passive reference off before analysing data.
What troubleshooting is recommended if efficiency of amplification is reduced with standard dilutions?
It has been reported that efficiency can decrease with subsequent dilutions for the standard curve. We recommend avoiding this by diluting the standards in 10 mM Tris-HCl pH 8.0, 0.1 mM EDTA, 0.05% Tween-20. EDTA is a chelating agent and it plays a role in preventing DNAse activity1. Tween-20 is a detergent and prevents the DNA from adsorbing to the sides of the tubes2. Most microcentrifuges are made of polypropylene and research has demonstrated that DNA sticks very well to polypropylene3.
Standards should not be frozen after diluting them. Even in the presence of detergent, freezing seems to cause DNA to bind irreversibly to polypropylene. We suggest leaving your standards at 4°C and preparing a fresh batch every few weeks.
1 Barra, G. B. et al. EDTA-mediated inhibition of DNases protects circulating cell-free DNA from ex vivo degradation in blood samples. Clin Biochem 48, 976-981, doi:10.1016/j.clinbiochem.2015.02.014 (2015).
2 Linnarsson, S. Recent advances in DNA sequencing methods – general principles of sample preparation. Exp Cell Res 316, 1339-1343, doi:10.1016/j.yexcr.2010.02.036 (2010).
3 Gaillard, C. & Strauss, F. Avoiding adsorption of DNA to polypropylene tubes and denaturation of short DNA fragments. Technical Tips Online 3, 3 (1998).
What troubleshooting is recommended if the reaction is being inhibited?
If inhibition is observed, the amount of template in the reaction can be decreased. This will increase the Ct value but lower the likelihood of inhibitors interfering with the Taq DNA polymerase activity. If this doesn’t work, try adding 0.4-4 mg/ml of BSA to the reaction1,2. Ensure the cycling conditions in our product manual are adhered to.
1 Kreader, C. A. Relief of amplification inhibition in PCR with bovine serum albumin or T4 gene 32 protein. Appl Environ Microbiol 62, 1102-1106 (1996).
2 Wilson, I. G. Inhibition and facilitation of nucleic acid amplification. Appl Environ Microbiol 63, 3741-3751 (1997).
What troubleshooting is recommended if there are non-specific products using qPCRBIO Probe Mixes?
There are different options to consider when optimising the reaction:
- Reduce the annealing/extension time to 5 seconds
- Increase the annealing/extension temperature from 60 to 65°C
- Dilute the DNA template by starting with 5ng of DNA and using a 10x template dilution series. In addition to running these on a gel to see if the non-specific products persist, the efficiency of the reaction can be calculated with the software of the qPCR instrument after doing the template dilution. If the efficiency is between 90 – 110%, then the amplicon is being doubled every cycle.
What troubleshooting is recommended when Ct values are higher than usual?
Higher Ct values are generally indicative of delayed amplification. This could most likely be due to an excess of template in the reaction resulting in the primers and probes being bound on different DNA molecules. Samples usually have a lot of DNA other than the target gene and this can scatter the oligos. We recommend diluting the samples (10x-1000x) to solve this.
Moreover, annealing/extension temperature can also be increased to make the binding of the oligos more specific to the target sequence and decrease background signal.
When performing a multiplex, what is the recommended concentration of each primer?
We recommend using 0.4 µM of each primer. There is a degree of flexibility around this recommended concentration however, the primer concentration should not be increased, as this may significantly affect the activity of the enzyme.
Why is there a non-specific product when using the same primers and PCR conditions as a competitor’s product?
It’s most likely because the time for the 1st step (hot start) is too short. Ensure that the hot start phase is done at 95°C for 2 minutes to fully activate the enzyme. The recommended thermal profile is:
- 95°C (120 seconds)
- 40 cycles: 95°C (5-15 seconds) – 60°C (20-30 seconds)
- Melt
If non-specific products are still obtained, we recommend raising the annealing/extension temperature from 60°C to 65°C, depending on the primer set used.
More Information
qPCRBIO Probe Mix is the robust choice for all your probe-based real-time PCR needs. Our universal kit can be used to reliably detect extremely low copy number targets and quantify any DNA template including genomic, cDNA and viral sequences.
By combining antibody-mediated hot start technology with the latest advances in buffer chemistry, we offer market-leading performance with minimal or no optimisation required. The enhanced sensitivity makes qPCRBIO Probe Mix the perfect choice for multiplexing.
Use our qPCR Selection Tool to find out which ROX variant is compatible with your instrument.
Applications
- Absolute quantification
- Relative gene expression analysis
- TaqMan®, Scorpions® and molecular beacon probes
- Detection of extremely low copy number targets
- Diagnostic real-time PCR
- Genotyping & allelic discrimination
Specifications
qPCRBIO Probe Mix Lo-ROX
Component
100 Reactions
500 Reactions
2000 Reactions
5000 Reactions
5000 Reactions
2x qPCRBIO Probe Mix Lo-ROX
1 x 1mL
5 x 1mL
20 x 1mL
1 x 50mL Bottle
50 x 1mL in Pouch
qPCRBIO Probe Mix Hi-ROX
Component
100 Reactions
500 Reactions
2000 Reactions
5000 Reactions
5000 Reactions
2x qPCRBIO Probe Mix Hi-ROX
1 x 1mL
5 x 1mL
20 x 1mL
1 x 50mL Bottle
50 x 1mL in Pouch
qPCRBIO Probe Mix No-ROX
Component
100 Reactions
500 Reactions
2000 Reactions
5000 Reactions
5000 Reactions
2x qPCRBIO Probe Mix No-ROX
1 x 1mL
5 x 1mL
20 x 1mL
1 x 50mL Bottle
50 x 1mL in Pouch
qPCRBIO Probe Mix Separate-ROX
Component
100 Reactions
500 Reactions
2000 Reactions
5000 Reactions
5000 Reactions
2x qPCRBIO Probe Mix No-ROX
1 x 1mL
5 x 1mL
20 x 1mL
1 x 50mL Bottle
50 x 1mL in Pouch
50μM ROX Additive
1 x 200μL
1 x 200μL
4 x 200μL
2 x 520μL
2 x 520μL
qPCRBIO Probe Mix Lo-ROX
Component
2x qPCRBIO Probe Mix Lo-ROX
100 Reactions
1 x 1mL
500 Reactions
5 x 1mL
2000 Reactions
20 x 1mL
5000 Reactions
1 x 50mL Bottle
5000 Reactions
50 x 1mL in Pouch
qPCRBIO Probe Mix Hi-ROX
Component
2x qPCRBIO Probe Mix Hi-ROX
100 Reactions
1 x 1mL
500 Reactions
5 x 1mL
2000 Reactions
20 x 1mL
5000 Reactions
1 x 50mL Bottle
5000 Reactions
50 x 1mL in Pouch
qPCRBIO Probe Mix No-ROX
Component
2x qPCRBIO Probe Mix No-ROX
100 Reactions
1 x 1mL
500 Reactions
5 x 1mL
2000 Reactions
20 x 1mL
5000 Reactions
1 x 50mL Bottle
5000 Reactions
50 x 1mL in Pouch
qPCRBIO Probe Mix Separate-ROX
Component
2x qPCRBIO Probe Mix No-ROX
50μM ROX Additive
100 Reactions
1 x 1mL
1 x 200μL
500 Reactions
5 x 1mL
1 x 200μL
2000 Reactions
20 x 1mL
4 x 200μL
5000 Reactions
1 x 50mL Bottle
2 x 520μL
5000 Reactions
50 x 1mL in Pouch
2 x 520μL
Reaction Volume
Storage
20μL
On arrival, products should be stored between -30 and -15°C. If stored correctly the kit will retain full activity for 12 months.
Reaction Volume
20μL
Storage
On arrival, products should be stored between -30 and -15°C. If stored correctly the kit will retain full activity for 12 months.
Instrument Compatibility
This product is compatible with all standard and fast cycling qPCR instruments. Use our qPCR Selection Tool to find out which ROX variant is compatible with your instrument.
Documents
Product Flyers
Product Manuals
Material Safety Data Sheets
Certificate of Analysis Finder
FAQs
Can products generated with qPCRBIO Probe Mixes be digested, cloned, and sequenced?
Yes, PCR products generated with qPCRBIO Probe Mixes have the same characteristics as PCR products generated with wild-type Taq polymerase. They may be sequenced or digested with restriction endonucleases using standard protocols. Products are 3′-d(A)-tailed and may be used for TA cloning or may be blunt-ended or digested with restriction enzymes prior to cloning. For best results, we recommend purifying the PCR products using any standard PCR clean-up kit.
Can ROX have a negative impact on the reaction?
ROX (6-carboxy-X-rhodamine) is used as a passive reference dye in ROX-dependent real-time PCR instruments to normalize for variations of fluorescence levels that can arise mainly due to optical path variations among wells. Normalisation of the fluorescence intensity (Rn) is done in real-time PCR software by dividing the emission intensity of the specific signal by the emission intensity of ROX.
ROX does not take part in the PCR reaction and its fluorescence levels are not proportional to the quantity of DNA in each well, so the addition of this fluorophore to a mix provides a constant fluorescent signal during amplification.
Different types of real-time PCR instruments requiring a passive reference standard have different optimal concentrations of ROX, mainly due to the different optical configurations of each system (i.e. the different type of excitation source and optics used).
The addition of either too little or too much ROX would result in a very noisy signal impacting on the results of the reaction. Therefore, it is extremely important for the user to:
- Determine the correct ROX concentration to optimise real-time PCR results, and
- Check the ROX settings on the software used to set up the reaction
A useful selection tool for the most commonly used systems can be found here.
Can the activation step at 95°C be increased to 15 minutes if DNA needs to be isolated from bacteria (colony PCR) or from viral particles?
Yes. The usual activation time of 2 minutes is not a long enough time to ensure release of the DNA from the bacteria or virus sample. The activation step can be increased to 15 minutes without affecting the reaction yield.
Do qPCRBIO Probe Mixes contain the FAM dye in it?
No. Apart from ROX, there is no other dye in our mixes. You can therefore use any fluorophore-conjugated probe for your reaction. Keep in mind that if you’re using qPCRBIO Probe Blue Mixes, refer to Table 1 in the respective product manuals to ensure the blue dye will not interfere with the fluorescence of your fluorophore.
Is it normal if the fluorescence of qPCRBIO Probe Mixes differs from the one obtained with competitors’ products?
Different products could give a different plateau of fluorescence. However, this has no impact on quantification accuracy and Ct values will not differ among products.
Is the storage of sample DNA in 1x TE (10 mM Tris-HCl / 1 mM EDTA) buffer compatible with subsequent qPCR using qPCRBIO Probe Mixes?
Yes, this storage buffer is compatible. The EDTA will chelate some of the magnesium in the mix, but not significantly enough to affect the reaction.
What are the ROX concentrations in qPCRBIO Probe Mixes?
The qPCRBIO Probe Mixes that contain passive reference dyes come in different formulations, each with a different concentration of the passive reference dye:
- qPCRBIO Probe Mix Lo-ROX (PB20.21) contains 112 nM ROX.
- qPCRBIO Probe Mix Hi-ROX (PB20.22) contains 1.12 µM ROX.
- qPCRBIO Probe Mix No-ROX (PB20.23) does not contain ROX.
- qPCRBIO Probe Mix Separate-ROX (PB20.24) 2x mix contains no ROX and include a separate tube of 50 µM ROX additive. This enables you to choose what concentration of ROX you’d like to use.
You can use our qPCR Selection Tool under the Resources drop-down menu to determine which of our mixes are best suited for your qPCR machine.
What is included in qPCRBIO Probe Mixes?
qPCRBIO Probe Mixes are ready to use qPCR 2x Mastermixes. You only need to add primers, template DNA and PCR grade water during reaction set up.
What is ROX?
ROX is a passive reference dye which means it does not take part in the PCR reaction. It is used to normalise non-PCR related fluctuations in fluorescence.
What is the concentration of dNTPs in qPCRBIO Probe Mixes?
All qPCRBIO Probe Mixes contain dNTPs at a concentration of 2 mM (0.5 mM each). This means the final concentration in the reaction is 1 mM (0.25 mM each).
What is the MgCl2 concentration in qPCRBIO Probe Mixes?
All qPCRBIO Probe Mixes contain MgCl2 at a concentration of 12 mM. This means the final concentration in the reaction is 6 mM.
What should be considered if the normalisation of signal with ROX appears to be lower relative to a competitor’s mix?
Ensure you are using the right concentration of ROX because different instruments require different ROX concentrations. For example, if a qPCRBIO Probe Mix Hi-ROX is used with an instrument requiring a Lo-ROX mix, the software will normalise the qPCRBIO signal against the Hi-ROX level. This will significantly reduce the fluorescence level of the qPCRBIO mix relative to competitors’ mixes, if in that case the Lo-ROX mix was chosen for the competitor.
When comparing mixes from different manufacturers, it is better to carry out separate runs or turn the passive reference off before analysing data.
What troubleshooting is recommended if efficiency of amplification is reduced with standard dilutions?
It has been reported that efficiency can decrease with subsequent dilutions for the standard curve. We recommend avoiding this by diluting the standards in 10 mM Tris-HCl pH 8.0, 0.1 mM EDTA, 0.05% Tween-20. EDTA is a chelating agent and it plays a role in preventing DNAse activity1. Tween-20 is a detergent and prevents the DNA from adsorbing to the sides of the tubes2. Most microcentrifuges are made of polypropylene and research has demonstrated that DNA sticks very well to polypropylene3.
Standards should not be frozen after diluting them. Even in the presence of detergent, freezing seems to cause DNA to bind irreversibly to polypropylene. We suggest leaving your standards at 4°C and preparing a fresh batch every few weeks.
1 Barra, G. B. et al. EDTA-mediated inhibition of DNases protects circulating cell-free DNA from ex vivo degradation in blood samples. Clin Biochem 48, 976-981, doi:10.1016/j.clinbiochem.2015.02.014 (2015).
2 Linnarsson, S. Recent advances in DNA sequencing methods – general principles of sample preparation. Exp Cell Res 316, 1339-1343, doi:10.1016/j.yexcr.2010.02.036 (2010).
3 Gaillard, C. & Strauss, F. Avoiding adsorption of DNA to polypropylene tubes and denaturation of short DNA fragments. Technical Tips Online 3, 3 (1998).
What troubleshooting is recommended if the reaction is being inhibited?
If inhibition is observed, the amount of template in the reaction can be decreased. This will increase the Ct value but lower the likelihood of inhibitors interfering with the Taq DNA polymerase activity. If this doesn’t work, try adding 0.4-4 mg/ml of BSA to the reaction1,2. Ensure the cycling conditions in our product manual are adhered to.
1 Kreader, C. A. Relief of amplification inhibition in PCR with bovine serum albumin or T4 gene 32 protein. Appl Environ Microbiol 62, 1102-1106 (1996).
2 Wilson, I. G. Inhibition and facilitation of nucleic acid amplification. Appl Environ Microbiol 63, 3741-3751 (1997).
What troubleshooting is recommended if there are non-specific products using qPCRBIO Probe Mixes?
There are different options to consider when optimising the reaction:
- Reduce the annealing/extension time to 5 seconds
- Increase the annealing/extension temperature from 60 to 65°C
- Dilute the DNA template by starting with 5ng of DNA and using a 10x template dilution series. In addition to running these on a gel to see if the non-specific products persist, the efficiency of the reaction can be calculated with the software of the qPCR instrument after doing the template dilution. If the efficiency is between 90 – 110%, then the amplicon is being doubled every cycle.
What troubleshooting is recommended when Ct values are higher than usual?
Higher Ct values are generally indicative of delayed amplification. This could most likely be due to an excess of template in the reaction resulting in the primers and probes being bound on different DNA molecules. Samples usually have a lot of DNA other than the target gene and this can scatter the oligos. We recommend diluting the samples (10x-1000x) to solve this.
Moreover, annealing/extension temperature can also be increased to make the binding of the oligos more specific to the target sequence and decrease background signal.
When performing a multiplex, what is the recommended concentration of each primer?
We recommend using 0.4 µM of each primer. There is a degree of flexibility around this recommended concentration however, the primer concentration should not be increased, as this may significantly affect the activity of the enzyme.
Why is there a non-specific product when using the same primers and PCR conditions as a competitor’s product?
It’s most likely because the time for the 1st step (hot start) is too short. Ensure that the hot start phase is done at 95°C for 2 minutes to fully activate the enzyme. The recommended thermal profile is:
- 95°C (120 seconds)
- 40 cycles: 95°C (5-15 seconds) – 60°C (20-30 seconds)
- Melt
If non-specific products are still obtained, we recommend raising the annealing/extension temperature from 60°C to 65°C, depending on the primer set used.