To view your price please login or contact us
qPCRBIO SyGreen 1-Step Kits
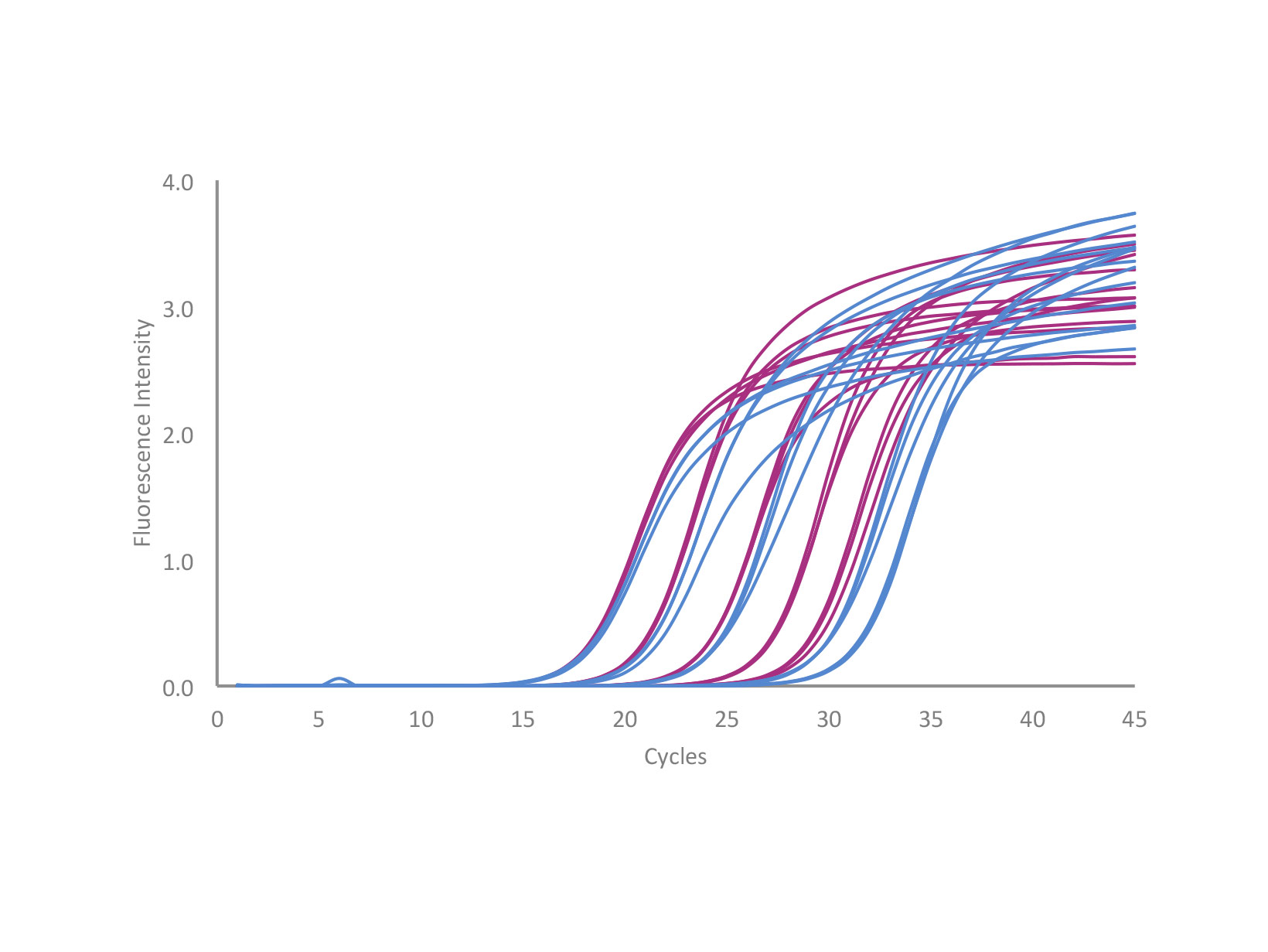
qPCRBIO SyGreen 1-Step Kits have been designed for fast, highly specific and ultra-sensitive cDNA synthesis and real-time PCR in a single tube.
Select from qPCRBIO SyGreen 1-Step Detect for the detection of extremely low copy number targets, or qPCRBIO SyGreen 1-Step Go for rapid and accurate results from high RNA template concentrations.
Features
- Thermostable reverse transcriptase 45°C to 55°C
- Advanced RNase inhibitor
- Non-PCR inhibiting intercalating dye
- Rapid extension rate for early Ct values
- Market-leading sensitivity
- Increased limit of detection
- Antibody-mediated hot start PCR
- Compatible on all standard and fast cycling real-time PCR platforms
More Information
qPCRBIO SyGreen 1-Step Kits have been designed for fast, highly specific and ultra-sensitive real-time RT-PCR. We use the latest developments in reverse transcriptase technology and buffer chemistry to give efficient cDNA synthesis and real-time PCR in a single tube.
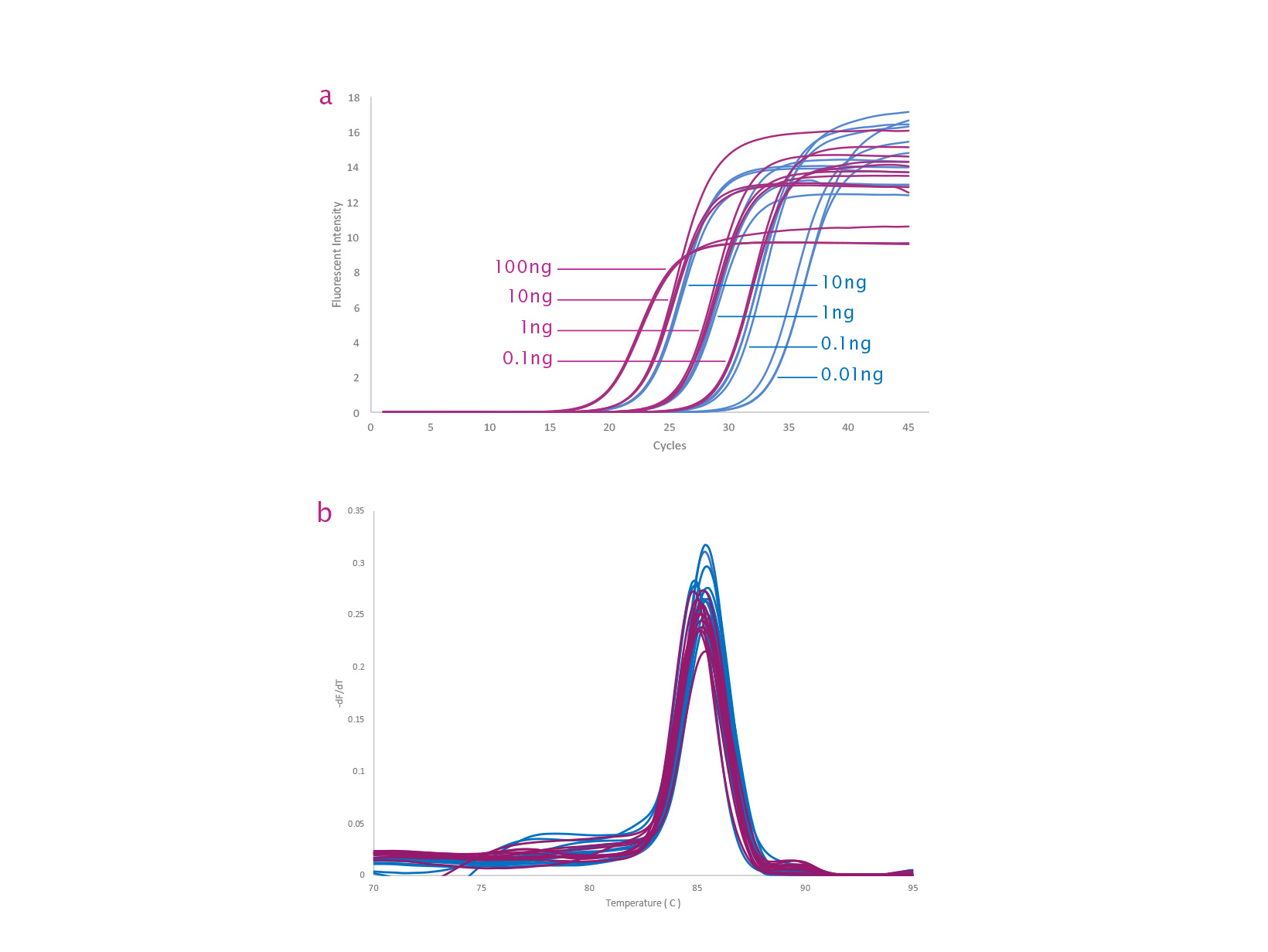
qPCRBIO SyGreen 1-Step Kits can be used to quantify any RNA template including mRNA, total RNA and viral RNA sequences. qPCRBIO SyGreen 1-Step Detect is designed for sensitivity and is ideally suited to the detection of extremely low copy number targets. qPCRBIO SyGreen 1-Step Go gives the earliest Ct and is formulated for rapid and accurate results from high template concentrations.
The kits include a thermostable and extremely active modified MMLV reverse transcriptase and advanced RNase inhibitor that prevents degradation of RNA by contaminating RNase. PCR amplification is powered by antibody-mediated hot start technology, preventing the formation of primer dimers and non-specific products leading to improved reaction sensitivity and specificity. Combining the latest developments in polymerase technology and advanced buffer chemistry we offer market- leading performance with minimal or no optimisation.
Applications
- Absolute quantification
- Relative gene expression analysis
- Detection of extremely low copy number targets
- qPCRBIO SyGreen 1-Step Detect recommended for template amounts of 1pg - 10ng total RNA or >0.01pg mRNA per reaction
- qPCRBIO SyGreen 1-Step Go recommended for template amounts of 10pg - 100ng total RNA or >0.01pg mRNA per reaction
Specifications
qPCRBIO SyGreen 1-Step Detect Lo-ROX
Component
100 Reactions
300 Reactions
1200 Reactions
2x qPCRBIO SyGreen 1-Step Lo-ROX
1 x 1mL
3 x 1mL
12 x 1mL
20x RTase with RNase Inhibitor
1 x 200μL
3 x 200μL
12 x 200μL
qPCRBIO SyGreen 1-Step Detect Hi-ROX
Component
100 Reactions
300 Reactions
1200 Reactions
2x qPCRBIO SyGreen 1-Step Hi-ROX
1 x 1mL
3 x 1mL
12 x 1mL
20x RTase with RNase Inhibitor
1 x 200μL
3 x 200μL
12 x 200μL
qPCRBIO SyGreen 1-Step Go Lo-ROX
Component
100 Reactions
300 Reactions
1200 Reactions
2x qPCRBIO SyGreen 1-Step Lo-ROX
1 x 1mL
3 x 1mL
12 x 1mL
20x RTase Go with RNase Inhibitor
1 x 100μL
3 x 100μL
12 x 100μL
qPCRBIO SyGreen 1-Step Go Hi-ROX
Component
100 Reactions
300 Reactions
1200 Reactions
2x qPCRBIO SyGreen 1-Step Hi-ROX
1 x 1mL
3 x 1mL
12 x 1mL
20x RTase Go with RNase Inhibitor
1 x 100μL
3 x 100μL
12 x 100μL
qPCRBIO SyGreen 1-Step Detect Lo-ROX
Component
2x qPCRBIO SyGreen 1-Step Lo-ROX
20x RTase with RNase Inhibitor
100 Reactions
1 x 1mL
1 x 200μL
300 Reactions
3 x 1mL
3 x 200μL
1200 Reactions
12 x 1mL
12 x 200μL
qPCRBIO SyGreen 1-Step Detect Hi-ROX
Component
2x qPCRBIO SyGreen 1-Step Hi-ROX
20x RTase with RNase Inhibitor
100 Reactions
1 x 1mL
1 x 200μL
300 Reactions
3 x 1mL
3 x 200μL
1200 Reactions
12 x 1mL
12 x 200μL
qPCRBIO SyGreen 1-Step Go Lo-ROX
Component
2x qPCRBIO SyGreen 1-Step Lo-ROX
20x RTase Go with RNase Inhibitor
100 Reactions
1 x 1mL
1 x 100μL
300 Reactions
3 x 1mL
3 x 100μL
1200 Reactions
12 x 1mL
12 x 100μL
qPCRBIO SyGreen 1-Step Go Hi-ROX
Component
2x qPCRBIO SyGreen 1-Step Hi-ROX
20x RTase Go with RNase Inhibitor
100 Reactions
1 x 1mL
1 x 100μL
300 Reactions
3 x 1mL
3 x 100μL
1200 Reactions
12 x 1mL
12 x 100μL
Reaction Volume
Storage
20μL
On arrival, products should be stored between -30 and -15°C. If stored correctly the kit will retain full activity for 12 months.
Reaction Volume
20μL
Storage
On arrival, products should be stored between -30 and -15°C. If stored correctly the kit will retain full activity for 12 months.
Instrument Compatibility
This product is compatible with all standard and fast cycling qPCR instruments. Use our qPCR Selection Tool to find out which ROX variant is compatible with your instrument.
Documents
Product Flyers
Product Manuals
Material Safety Data Sheets
Certificate of Analysis Finder
FAQs
Beyond what Ct value are results unreliable?
Ct values can vary between template concentrations, reaction optimisation, instruments and laboratories so care must be taken when selecting a cut-off Ct value. Generally, Ct values over 35 would begin to be considered unreliable. However, late Cts could be observed for inefficient reactions using low template copy number. It is always good practice to normalise cut-offs with relative or absolute quantification methods. It is also recommended to run and analyse melt curves or gels of the products to determine products of any late amplifications.
Can I use qPCRBIO SyGreen 1-Step Detect / Go for both one-step and two-step RT-PCR?
No, this kit will not work for 2-step reactions. For 2-step reactions, we recommend the qPCRBIO cDNA Synthesis Kit (PB30.11) or UltraScript 2.0 cDNA Synthesis Kit (PB30.31) for more difficult targets, along with real time PCR kits depending on your needs.
Can ROX have a negative impact on the reaction?
ROX (6-carboxy-X-rhodamine) is used as a passive reference dye in ROX-dependent real-time PCR instruments to normalize for variations of fluorescence levels that can arise mainly due to optical path variations among wells. Normalisation of the fluorescence intensity (Rn) is done in real-time PCR software by dividing the emission intensity of the specific signal by the emission intensity of ROX.
ROX does not take part in the PCR reaction and its fluorescence levels are not proportional to the quantity of DNA in each well, so the addition of this fluorophore to a mix provides a constant fluorescent signal during amplification.
Different types of real-time PCR instruments requiring a passive reference standard have different optimal concentrations of ROX, mainly due to the different optical configurations of each system (i.e. the different type of excitation source and optics used).
The addition of either too little or too much ROX would result in a very noisy signal impacting on the results of the reaction. Therefore, it is extremely important for the user to:
- Determine the correct ROX concentration to optimise real-time PCR results, and
- Check the ROX settings on the software used to set up the reaction
A useful selection tool for the most commonly used systems can be found here.
Can the activation time for the hot-start DNA polymerase be altered?
We recommend using a minimum of 2 minutes for activating the polymerase. Longer times of up to 15 minutes can also be used with no detrimental effects to the enzyme.
Do I need 1-step or 2-step reaction?
1-step
Both the cDNA synthesis and PCR reactions occur in the same mix. This option is suited for high throughput applications due to its speed and ease of set up. There is also a reduced risk of contamination. It is not ideal for low quality RNA samples or if the cDNA is required for archive or separate analysis.
2-step
The cDNA synthesis and PCR reactions occur separately. This option is better suited if the cDNA product needs to be retained for analysis. It also allows for higher levels of reaction optimisation. It permits control over the type and concentration of enzyme, RNA input and concentration of cDNA which, in turn, results in higher sensitivity compared to the 1-step format.
Do I need to use an RNase inhibitor in my RT reaction?
No, the RTase Go contains an RNase inhibitor to prevent any degradation and increase sensitivity.
General troubleshooting for low product/late Ct values:
If you are observing unusually late Ct values, try diluting the template RNA. By doing this, you are diluting any inhibitors which may be present to a concentration where they do not inhibit the reaction. Additionally, try increasing the reverse transcription step to 55 °C and increasing the annealing/extension temperatures. This may help resolve difficulties caused by secondary structures present in the RNA template and/or primers.
In cases where reaction inhibition may be involved, try reducing the amount of template1 or add 0.4 – 4.4 mg/ml BSA to the reaction2.
For more specific problems contact [email protected] with the following information:
- Amplicon size
- Reaction setup
- Cycling conditions
- Screen grabs of amplification traces and melting profile
1 Scipioni et al. A SYBR Green RT-PCR assay in single tube to detect human and bovine noroviruses and control for inhibition. Virology Journal.5:94 (2008). doi: 1186/1743-422X-5-94
2 Plante et al. The use of bovine serum albumin to improve the RT‐qPCR detection of foodborne viruses rinsed from vegetable surfaces. Applied Microbiology. 52:3 (2010) doi: https://doi.org/10.1111/j.1472-765X.2010.02989.x
What are the differences between probe and dye-based mixes?
Probe based kits such as qPCRBIO Probe 1-Step Go offer higher sensitivity and are unlikely to show non-template amplifications. Multiplexes can be measured using amplicons with different fluorophores for specific probes, which cannot be achieved using dyes.
Dye based systems such as qPCRBIO SyGreen 1-Step Detect | 1-Step Go detect any intact dsDNA and will therefore show primer dimers and off-target/non-template amplifications. These can be separated from product peaks by analysing melting curves.
What is ROX and do I need it?
ROX is a passive reference dye which means it does not take part in the PCR reaction. It is used to normalise non-PCR related fluctuations in fluorescence. You can use our qPCR Selection Tool under the Resources drop-down menu to determine which of our qPCR mixes are best suited for your qPCR machine.
What is the difference between qPCRBIO SyGreen 1-Step Go and qPCRBIO SyGreen 1-Step Detect?
qPCRBIO SyGreen 1-step Detect has been engineered to have high sensitivity for rapid and accurate results from low template concentrations 1pg of total RNA or 0.01pg of mRNA. qPCRBIO SyGreen 1-step Go has been developed for rapid (early Ct) and accurate results from high template concentrations from 10pg total RNA or 0.01pg of mRNA.
What should I consider if I want to compare qPCRBIO SyGreen 1-Step kit with an alternative manufacturer?
It is important to use what the manufacturer prescribes in the product manual with respect to PCR set up and cycling conditions. It’s also a good idea to use multiple positive controls and to amplify against a template dilution series.
What’s the difference between PCRBIO 1-Step Go RT-PCR Kit, qPCRBIO Probe 1-Step Go and qPCRBIO SyGreen 1-Step Detect|Go Kits?
The PCRBIO 1-step Go RT-PCR Kit has been developed for endpoint RT-PCR. The qPCRBIO Probe 1-Step Go and qPCRBIO SyGreen 1-Step Detect|Go are our probe- and dye-based options for real-time RT-PCR, respectively.
Will this work for micro RNA templates?
Yes, qPCRBIO SyGreen 1-Step Detect | Go can be used for micro RNA templates. Although we do no sell dedicated kits, all of our RTases can be used for miRNA quantification and analysis.
We advise that you use one of the two following approaches:
- Use universal RT primers and add poly(A) or poly(U) tails (e.g. by poly(U)-polymerase), followed by cDNA synthesis using universal primers1,2.
- Use specific RT primers and omit the tailing step1,3-5.
If you are unfamiliar with the specifics of those approaches, please refer to the reference list below, which serve as a guideline.
1 Dave, V. P. et al. MicroRNA amplification and detection technologies: opportunities and challenges for point of care diagnostics. Lab Invest 99, 452-469, doi:10.1038/s41374-018-0143-3 (2019).
2 Mei, Q. et al. A facile and specific assay for quantifying microRNA by an optimized RT-qPCR approach. PLoS One 7, e46890, doi:10.1371/journal.pone.0046890 (2012).
3 Chen, C. et al. Real-time quantification of microRNAs by stem-loop RT-PCR. Nucleic Acids Res 33, e179, doi:10.1093/nar/gni178 (2005).
4 Raymond, C. K., Roberts, B. S., Garrett-Engele, P., Lim, L. P. & Johnson, J. M. Simple, quantitative primer-extension PCR assay for direct monitoring of microRNAs and short-interfering RNAs. RNA 11, 1737-1744, doi:10.1261/rna.2148705 (2005).
5 Androvic, P., Valihrach, L., Elling, J., Sjoback, R. & Kubista, M. Two-tailed RT-qPCR: a novel method for highly accurate miRNA quantification. Nucleic Acids Res 45, e144, doi:10.1093/nar/gkx588 (2017).
More Information
qPCRBIO SyGreen 1-Step Kits have been designed for fast, highly specific and ultra-sensitive real-time RT-PCR. We use the latest developments in reverse transcriptase technology and buffer chemistry to give efficient cDNA synthesis and real-time PCR in a single tube.
qPCRBIO SyGreen 1-Step Kits can be used to quantify any RNA template including mRNA, total RNA and viral RNA sequences. qPCRBIO SyGreen 1-Step Detect is designed for sensitivity and is ideally suited to the detection of extremely low copy number targets. qPCRBIO SyGreen 1-Step Go gives the earliest Ct and is formulated for rapid and accurate results from high template concentrations.
The kits include a thermostable and extremely active modified MMLV reverse transcriptase and advanced RNase inhibitor that prevents degradation of RNA by contaminating RNase. PCR amplification is powered by antibody-mediated hot start technology, preventing the formation of primer dimers and non-specific products leading to improved reaction sensitivity and specificity. Combining the latest developments in polymerase technology and advanced buffer chemistry we offer market- leading performance with minimal or no optimisation.
Applications
- Absolute quantification
- Relative gene expression analysis
- Detection of extremely low copy number targets
- qPCRBIO SyGreen 1-Step Detect recommended for template amounts of 1pg - 10ng total RNA or >0.01pg mRNA per reaction
- qPCRBIO SyGreen 1-Step Go recommended for template amounts of 10pg - 100ng total RNA or >0.01pg mRNA per reaction
Specifications
qPCRBIO SyGreen 1-Step Detect Lo-ROX
Component
100 Reactions
300 Reactions
1200 Reactions
2x qPCRBIO SyGreen 1-Step Lo-ROX
1 x 1mL
3 x 1mL
12 x 1mL
20x RTase with RNase Inhibitor
1 x 200μL
3 x 200μL
12 x 200μL
qPCRBIO SyGreen 1-Step Detect Hi-ROX
Component
100 Reactions
300 Reactions
1200 Reactions
2x qPCRBIO SyGreen 1-Step Hi-ROX
1 x 1mL
3 x 1mL
12 x 1mL
20x RTase with RNase Inhibitor
1 x 200μL
3 x 200μL
12 x 200μL
qPCRBIO SyGreen 1-Step Go Lo-ROX
Component
100 Reactions
300 Reactions
1200 Reactions
2x qPCRBIO SyGreen 1-Step Lo-ROX
1 x 1mL
3 x 1mL
12 x 1mL
20x RTase Go with RNase Inhibitor
1 x 100μL
3 x 100μL
12 x 100μL
qPCRBIO SyGreen 1-Step Go Hi-ROX
Component
100 Reactions
300 Reactions
1200 Reactions
2x qPCRBIO SyGreen 1-Step Hi-ROX
1 x 1mL
3 x 1mL
12 x 1mL
20x RTase Go with RNase Inhibitor
1 x 100μL
3 x 100μL
12 x 100μL
qPCRBIO SyGreen 1-Step Detect Lo-ROX
Component
2x qPCRBIO SyGreen 1-Step Lo-ROX
20x RTase with RNase Inhibitor
100 Reactions
1 x 1mL
1 x 200μL
300 Reactions
3 x 1mL
3 x 200μL
1200 Reactions
12 x 1mL
12 x 200μL
qPCRBIO SyGreen 1-Step Detect Hi-ROX
Component
2x qPCRBIO SyGreen 1-Step Hi-ROX
20x RTase with RNase Inhibitor
100 Reactions
1 x 1mL
1 x 200μL
300 Reactions
3 x 1mL
3 x 200μL
1200 Reactions
12 x 1mL
12 x 200μL
qPCRBIO SyGreen 1-Step Go Lo-ROX
Component
2x qPCRBIO SyGreen 1-Step Lo-ROX
20x RTase Go with RNase Inhibitor
100 Reactions
1 x 1mL
1 x 100μL
300 Reactions
3 x 1mL
3 x 100μL
1200 Reactions
12 x 1mL
12 x 100μL
qPCRBIO SyGreen 1-Step Go Hi-ROX
Component
2x qPCRBIO SyGreen 1-Step Hi-ROX
20x RTase Go with RNase Inhibitor
100 Reactions
1 x 1mL
1 x 100μL
300 Reactions
3 x 1mL
3 x 100μL
1200 Reactions
12 x 1mL
12 x 100μL
Reaction Volume
Storage
20μL
On arrival, products should be stored between -30 and -15°C. If stored correctly the kit will retain full activity for 12 months.
Reaction Volume
20μL
Storage
On arrival, products should be stored between -30 and -15°C. If stored correctly the kit will retain full activity for 12 months.
Instrument Compatibility
This product is compatible with all standard and fast cycling qPCR instruments. Use our qPCR Selection Tool to find out which ROX variant is compatible with your instrument.
Documents
Product Flyers
Product Manuals
Material Safety Data Sheets
Certificate of Analysis Finder
FAQs
Beyond what Ct value are results unreliable?
Ct values can vary between template concentrations, reaction optimisation, instruments and laboratories so care must be taken when selecting a cut-off Ct value. Generally, Ct values over 35 would begin to be considered unreliable. However, late Cts could be observed for inefficient reactions using low template copy number. It is always good practice to normalise cut-offs with relative or absolute quantification methods. It is also recommended to run and analyse melt curves or gels of the products to determine products of any late amplifications.
Can I use qPCRBIO SyGreen 1-Step Detect / Go for both one-step and two-step RT-PCR?
No, this kit will not work for 2-step reactions. For 2-step reactions, we recommend the qPCRBIO cDNA Synthesis Kit (PB30.11) or UltraScript 2.0 cDNA Synthesis Kit (PB30.31) for more difficult targets, along with real time PCR kits depending on your needs.
Can ROX have a negative impact on the reaction?
ROX (6-carboxy-X-rhodamine) is used as a passive reference dye in ROX-dependent real-time PCR instruments to normalize for variations of fluorescence levels that can arise mainly due to optical path variations among wells. Normalisation of the fluorescence intensity (Rn) is done in real-time PCR software by dividing the emission intensity of the specific signal by the emission intensity of ROX.
ROX does not take part in the PCR reaction and its fluorescence levels are not proportional to the quantity of DNA in each well, so the addition of this fluorophore to a mix provides a constant fluorescent signal during amplification.
Different types of real-time PCR instruments requiring a passive reference standard have different optimal concentrations of ROX, mainly due to the different optical configurations of each system (i.e. the different type of excitation source and optics used).
The addition of either too little or too much ROX would result in a very noisy signal impacting on the results of the reaction. Therefore, it is extremely important for the user to:
- Determine the correct ROX concentration to optimise real-time PCR results, and
- Check the ROX settings on the software used to set up the reaction
A useful selection tool for the most commonly used systems can be found here.
Can the activation time for the hot-start DNA polymerase be altered?
We recommend using a minimum of 2 minutes for activating the polymerase. Longer times of up to 15 minutes can also be used with no detrimental effects to the enzyme.
Do I need 1-step or 2-step reaction?
1-step
Both the cDNA synthesis and PCR reactions occur in the same mix. This option is suited for high throughput applications due to its speed and ease of set up. There is also a reduced risk of contamination. It is not ideal for low quality RNA samples or if the cDNA is required for archive or separate analysis.
2-step
The cDNA synthesis and PCR reactions occur separately. This option is better suited if the cDNA product needs to be retained for analysis. It also allows for higher levels of reaction optimisation. It permits control over the type and concentration of enzyme, RNA input and concentration of cDNA which, in turn, results in higher sensitivity compared to the 1-step format.
Do I need to use an RNase inhibitor in my RT reaction?
No, the RTase Go contains an RNase inhibitor to prevent any degradation and increase sensitivity.
General troubleshooting for low product/late Ct values:
If you are observing unusually late Ct values, try diluting the template RNA. By doing this, you are diluting any inhibitors which may be present to a concentration where they do not inhibit the reaction. Additionally, try increasing the reverse transcription step to 55 °C and increasing the annealing/extension temperatures. This may help resolve difficulties caused by secondary structures present in the RNA template and/or primers.
In cases where reaction inhibition may be involved, try reducing the amount of template1 or add 0.4 – 4.4 mg/ml BSA to the reaction2.
For more specific problems contact [email protected] with the following information:
- Amplicon size
- Reaction setup
- Cycling conditions
- Screen grabs of amplification traces and melting profile
1 Scipioni et al. A SYBR Green RT-PCR assay in single tube to detect human and bovine noroviruses and control for inhibition. Virology Journal.5:94 (2008). doi: 1186/1743-422X-5-94
2 Plante et al. The use of bovine serum albumin to improve the RT‐qPCR detection of foodborne viruses rinsed from vegetable surfaces. Applied Microbiology. 52:3 (2010) doi: https://doi.org/10.1111/j.1472-765X.2010.02989.x
What are the differences between probe and dye-based mixes?
Probe based kits such as qPCRBIO Probe 1-Step Go offer higher sensitivity and are unlikely to show non-template amplifications. Multiplexes can be measured using amplicons with different fluorophores for specific probes, which cannot be achieved using dyes.
Dye based systems such as qPCRBIO SyGreen 1-Step Detect | 1-Step Go detect any intact dsDNA and will therefore show primer dimers and off-target/non-template amplifications. These can be separated from product peaks by analysing melting curves.
What is ROX and do I need it?
ROX is a passive reference dye which means it does not take part in the PCR reaction. It is used to normalise non-PCR related fluctuations in fluorescence. You can use our qPCR Selection Tool under the Resources drop-down menu to determine which of our qPCR mixes are best suited for your qPCR machine.
What is the difference between qPCRBIO SyGreen 1-Step Go and qPCRBIO SyGreen 1-Step Detect?
qPCRBIO SyGreen 1-step Detect has been engineered to have high sensitivity for rapid and accurate results from low template concentrations 1pg of total RNA or 0.01pg of mRNA. qPCRBIO SyGreen 1-step Go has been developed for rapid (early Ct) and accurate results from high template concentrations from 10pg total RNA or 0.01pg of mRNA.
What should I consider if I want to compare qPCRBIO SyGreen 1-Step kit with an alternative manufacturer?
It is important to use what the manufacturer prescribes in the product manual with respect to PCR set up and cycling conditions. It’s also a good idea to use multiple positive controls and to amplify against a template dilution series.
What’s the difference between PCRBIO 1-Step Go RT-PCR Kit, qPCRBIO Probe 1-Step Go and qPCRBIO SyGreen 1-Step Detect|Go Kits?
The PCRBIO 1-step Go RT-PCR Kit has been developed for endpoint RT-PCR. The qPCRBIO Probe 1-Step Go and qPCRBIO SyGreen 1-Step Detect|Go are our probe- and dye-based options for real-time RT-PCR, respectively.
Will this work for micro RNA templates?
Yes, qPCRBIO SyGreen 1-Step Detect | Go can be used for micro RNA templates. Although we do no sell dedicated kits, all of our RTases can be used for miRNA quantification and analysis.
We advise that you use one of the two following approaches:
- Use universal RT primers and add poly(A) or poly(U) tails (e.g. by poly(U)-polymerase), followed by cDNA synthesis using universal primers1,2.
- Use specific RT primers and omit the tailing step1,3-5.
If you are unfamiliar with the specifics of those approaches, please refer to the reference list below, which serve as a guideline.
1 Dave, V. P. et al. MicroRNA amplification and detection technologies: opportunities and challenges for point of care diagnostics. Lab Invest 99, 452-469, doi:10.1038/s41374-018-0143-3 (2019).
2 Mei, Q. et al. A facile and specific assay for quantifying microRNA by an optimized RT-qPCR approach. PLoS One 7, e46890, doi:10.1371/journal.pone.0046890 (2012).
3 Chen, C. et al. Real-time quantification of microRNAs by stem-loop RT-PCR. Nucleic Acids Res 33, e179, doi:10.1093/nar/gni178 (2005).
4 Raymond, C. K., Roberts, B. S., Garrett-Engele, P., Lim, L. P. & Johnson, J. M. Simple, quantitative primer-extension PCR assay for direct monitoring of microRNAs and short-interfering RNAs. RNA 11, 1737-1744, doi:10.1261/rna.2148705 (2005).
5 Androvic, P., Valihrach, L., Elling, J., Sjoback, R. & Kubista, M. Two-tailed RT-qPCR: a novel method for highly accurate miRNA quantification. Nucleic Acids Res 45, e144, doi:10.1093/nar/gkx588 (2017).