To view your price please login or contact us
PCRBIO Taq DNA Polymerase & Mixes
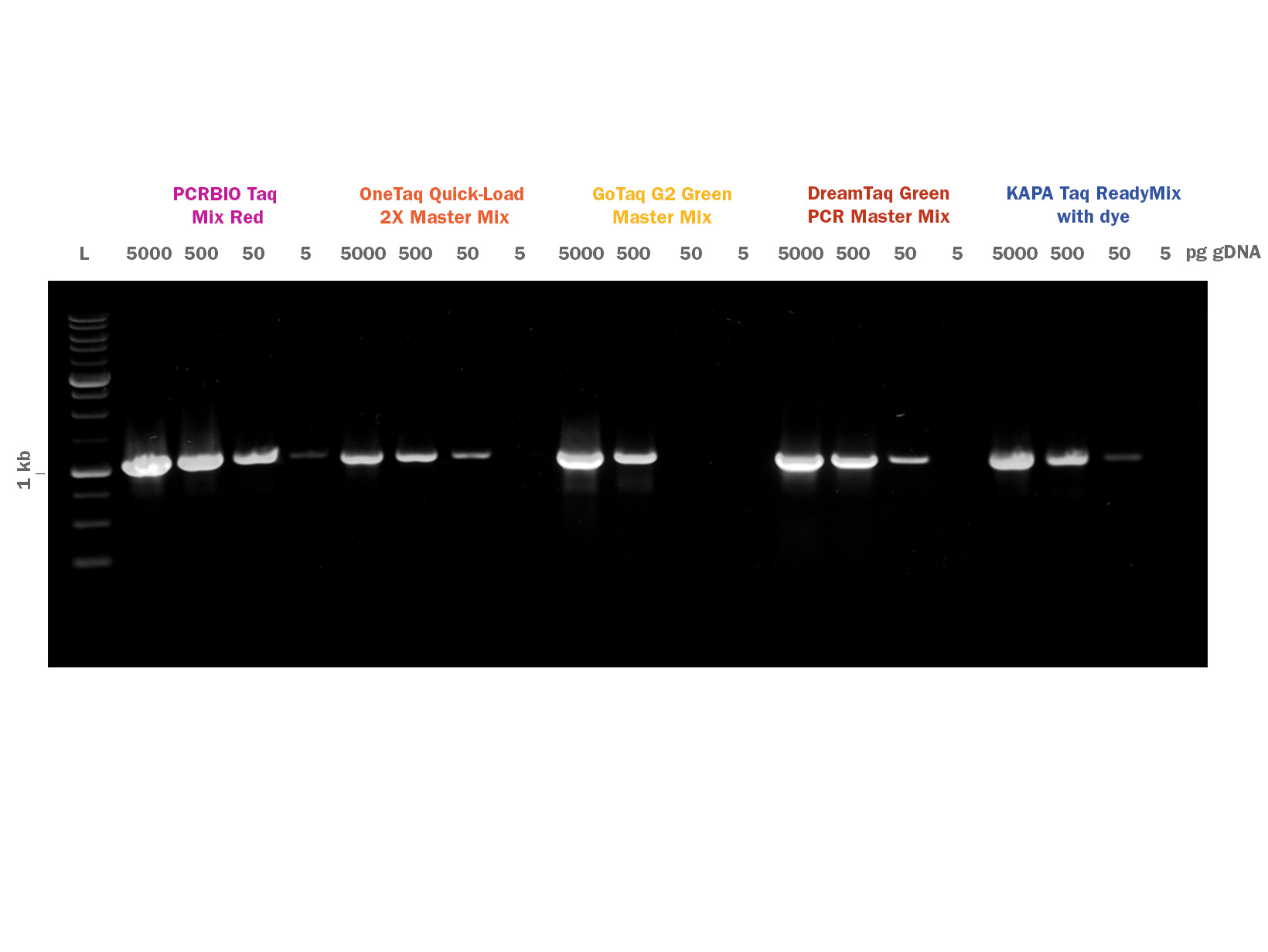
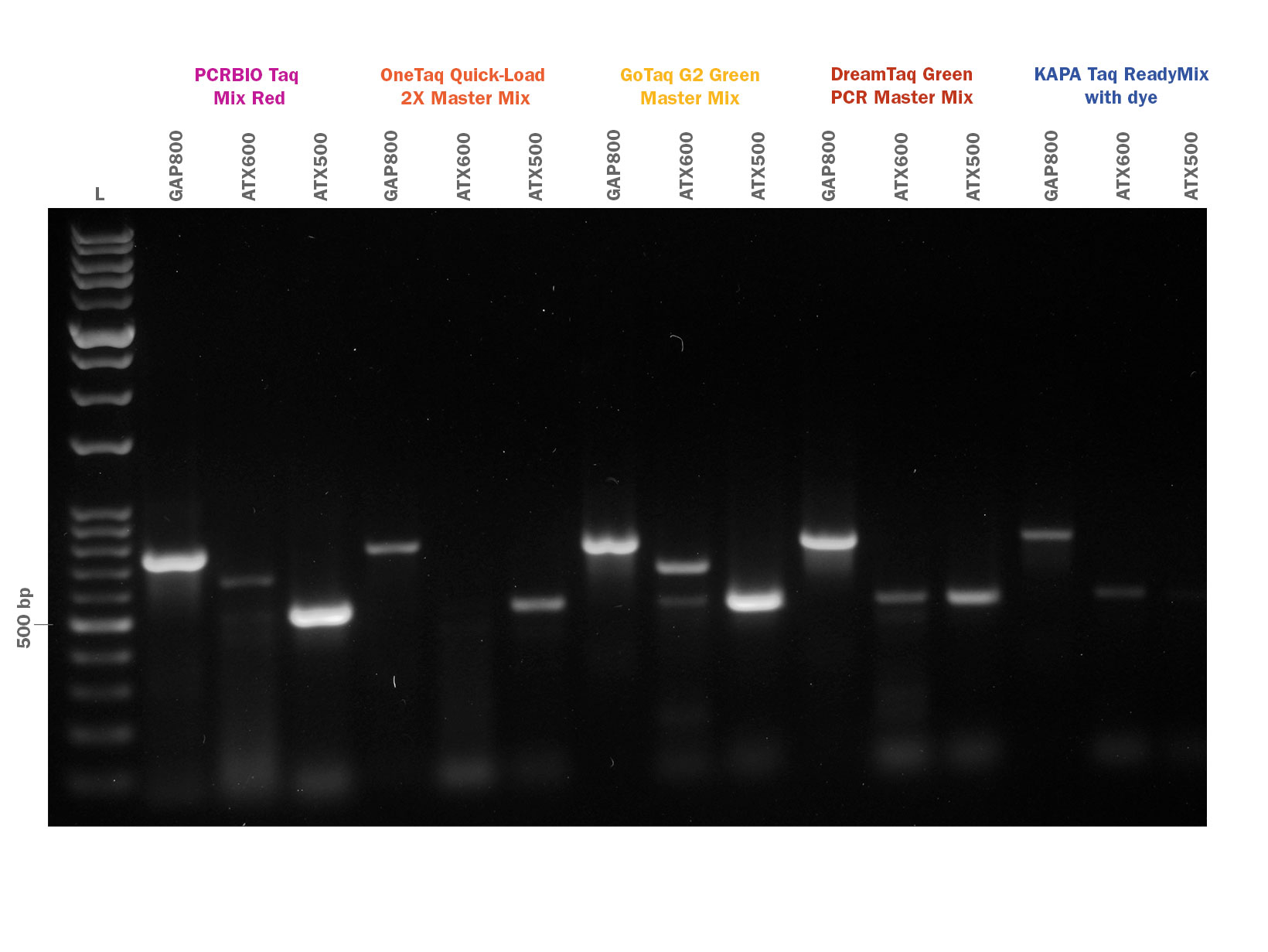
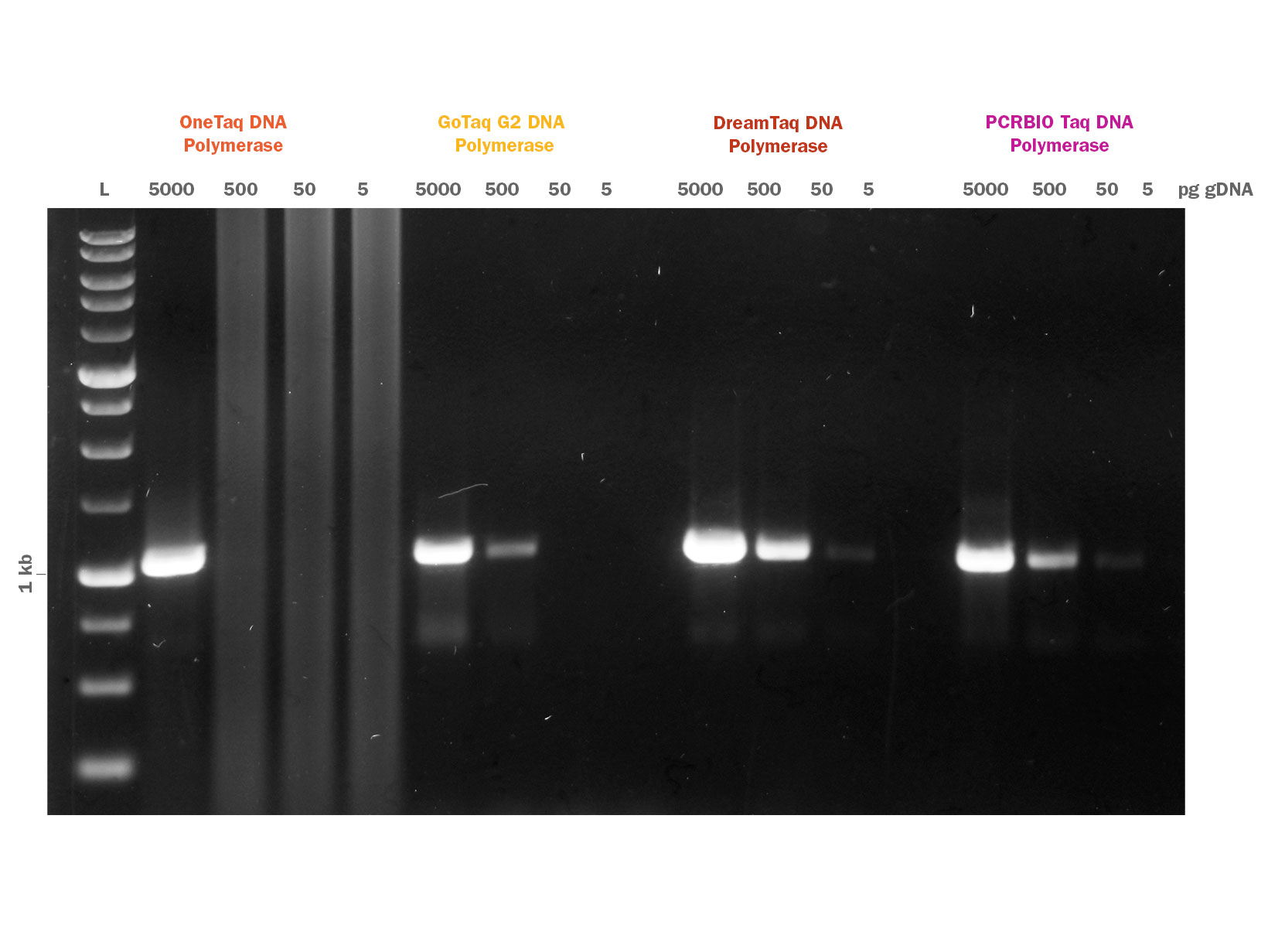
PCRBIO Taq DNA Polymerase is an affordable, versatile and robust enzyme for all your everyday PCR applications including genotyping, screening and library construction.
An enhanced 12-step purification strategy together with an optimised buffer system enable PCRBIO Taq DNA Polymerase to amplify with the highest speed, yield and specificity.
Features
- Increased PCR success rates with amplicons up to 6kb
- Ultra low background DNA
- Advanced buffer chemistry including Mg and dNTPs
- High yields under standard and fast PCR conditions
- Efficient specific amplification from complex templates including GC and AT-rich sequences
More Information
PCRBIO Taq DNA Polymerase is a robust enzyme for all your everyday PCR applications including genotyping, screening and library construction. PCRBIO Taq DNA Polymerase performs consistently well on a broad range of templates including both GC and AT rich.
PCRBIO Taq DNA Polymerase has 5’-3’ exonuclease activities, but no 3’-5’ exonuclease (proofreading) activity. The enzyme has the same error rate as wild-type taq DNA polymerase, approximately 1 error per 2.0 x 105 nucleotides incorporated.
PCRBIO Taq DNA Polymerase provides the research community with an affordable routine application polymerase that performs to the highest possible standard, with a versatility that allows you to amplify with the highest speed, yield, specificity and consistency on the market. PCR products generated are A-tailed and may be cloned into TA cloning vectors. The enzyme is room temperature stable for 4 weeks and is produced using an enhanced 12 step purification strategy which includes physical, chemical and enzymatic removal of host DNA.
For added convenience, PCRBIO Taq DNA Polymerase is available as a ready-to-use 2x mix containing all reaction components except primers and template. PCRBIO Taq Mix Red contains a red dye suitable for direct loading and tracking during agarose gel electrophoresis.
Applications
- Routine application PCR
- TA cloning
- High throughput PCR
- Methylated DNA
- Crude sample PCR
- Standard and fast PCR
- Efficient specific amplification from GC and AT-rich sequences
Specifications
PCRBIO Taq DNA Polymerase
Component
500 Units
2000 Units
4000 Units
PCRBIO Taq DNA Polymerase (5u/μL)
1 x 100μL
4 x 100μL
8 x 100μL
5x PCRBIO Reaction Buffer
4 x 1mL
16 x 1mL
32 x 1mL
PCRBIO Taq Mix
Component
200 Reactions
1000 Reactions
2x PCRBIO Taq Mix
5 x 1mL
25 x 1mL
PCRBIO Taq Mix Red
Component
200 Reactions
1000 Reactions
2x PCRBIO Taq Mix Red
5 x 1mL
25 x 1mL
PCRBIO Taq DNA Polymerase
Component
PCRBIO Taq DNA Polymerase (5u/μL)
5x PCRBIO Reaction Buffer
500 Units
1 x 100μL
4 x 1mL
2000 Units
4 x 100μL
16 x 1mL
4000 Units
8 x 100μL
32 x 1mL
PCRBIO Taq Mix
Component
2x PCRBIO Taq Mix
200 Reactions
5 x 1mL
1000 Reactions
25 x 1mL
PCRBIO Taq Mix Red
Component
2x PCRBIO Taq Mix Red
200 Reactions
5 x 1mL
1000 Reactions
25 x 1mL
Reaction Volume
Storage
50μL
On arrival, products should be stored between -30 and -15°C. If stored correctly the kit will retain full activity for 12 months.
Reaction Volume
50μL
Storage
On arrival, products should be stored between -30 and -15°C. If stored correctly the kit will retain full activity for 12 months.
Documents
Product Manuals
Material Safety Data Sheets
Certificate of Analysis Finder
FAQs
Can I use PCRBIO Taq DNA Polymerase if my assay requires a specialized buffer?
The 5x PCRBIO Reaction Buffer supplied with PCRBIO Taq DNA Polymerase has been developed specifically for this enzyme and we highly recommend using them together. However, PCRBIO Taq DNA Polymerase should be compatible with any PCR buffer developed for use with wild-type Taq. If you use a customised buffer with PCRBIO Taq DNA Polymerase, keep in mind reaction parameters such as annealing temperature and concentrations of the enzyme, template, dNTPs and MgCl2, may require optimisation.
Can PCRBIO Taq DNA Polymerase be used for colony PCR?
Yes. If you’re working from bacterial colonies use a sterile tip to pick a colony and re-suspend into the 50µl PCR reaction. If working from liquid culture add 5µl of overnight culture to the final mix. Follow the general protocol and increase the initial denaturation time to 10 min at 95°C.
Can PCRBIO Taq DNA Polymerase be used to amplify DNA from blood samples?
We recommend using PCRBIO HS Taq DNA Polymerase for amplifying DNA from blood samples. If you’d like to use PCRBIO Taq DNA Polymerase, use 2 µL blood sample to a 50 µL PCR reaction and follow the general protocol. Please note that blood components may inhibit the PCR reaction. Perform a serial dilution of the sample in order to find the optimal template concentration for the PCR amplification.
Can PCRBIO Taq DNA Polymerase proofread?
No. PCRBIO Taq DNA Polymerase has 5’-3’ exonuclease activities, but no 3’-5’ exonuclease (proofreading) activity.
Does PCRBIO Taq DNA Polymerase generate an A-tail or blunt end on my PCR product?
PCR products generated with PCRBIO Taq DNA Polymerase are A-tailed and this makes it suitable for cloning into TA vectors. For further reading, refer to this literature1.
1 Yao, S., Hart, D. J. & An, Y. Recent advances in universal TA cloning methods for use in function studies. Protein Eng Des Sel, doi:10.1093/protein/gzw047 (2016).
My results contain a high background of non-specific amplicons or smears. What trouble-shooting suggestions do you have?
If smears are a concern, it’s good practice to ensure they are not an artifact of running agarose gel electrophoresis with sub optimal conditions. Sub optimal conditions can include high voltage or not allowing enough time for the gel to set1.
You may also need to troubleshoot the PCR reaction and consider the suggestions below2.
- Primers should be designed to prevent primer-primer interactions and improve specificity.
- Increase the annealing temperature or conducting an annealing temperature gradient PCR to determine the optimal annealing temperature.
- Reduce the amount of template in the reaction. For high quality DNA, use 1–100 ng genomic DNA or ≤5 ng plasmid/lambda DNA per 50 µL reaction.
- Reduce the number of cycles.
- Reduce the amount of enzyme per reaction.
- Reduce the primer concentration, but not lower than 100 nM of each primer.
- Include DMSO in the reaction to a final concentration of 5%–10%.
1 Koontz, L. Agarose Gel Electrophoresis. Laboratory methods in enzymology : DNA. First edition. edn, Vol. 529 35-45 (2013).
2 Lorenz, T. C. Polymerase chain reaction: basic protocol plus troubleshooting and optimization strategies. J Vis Exp, e3998, doi:10.3791/3998 (2012).
My results show a very low yield. What trouble-shooting suggestions do you have?
You may want to consider the suggestions below and also refer to the literature1.
- Optimise the annealing temperature in an annealing temperature gradient PCR.
- Increase the amount of template in the reaction.
- Increase the number of cycles.
- Increase the amount of enzyme per reaction.
- Increase the primer concentration, but do not exceed 1 µM of each primer.
- Try a fresh dNTP solution.
- Optimise the MgCl2
1 Lorenz, T. C. Polymerase chain reaction: basic protocol plus troubleshooting and optimization strategies. J Vis Exp, e3998, doi:10.3791/3998 (2012).
What is the apparent Mw of the Red Mix dye on agarose gels?
This non-inhibitory dye, added to enable direct gel loading, runs at a similar rate to 200-300 bp DNA fragments on a 1% agarose gel and at 50-100 bp on a 2% agarose gel. You may notice a shift in this apparent molecular weight when running gels of different agarose content.
What is the difference between PCRBIO Taq DNA Polymerase and PCRBIO HS Taq DNA Polymerase?
PCRBIO HS Taq DNA Polymerase uses proprietary antibody-mediated hot start technology whereas PCRBIO Taq DNA Polymerase does not have this feature. The interaction of the antibody with the Taq DNA Polymerase leaves the enzyme inactive until the hot start step.
The hot start refers to the initial activation step at 95°C, which subsequently deactivates the antibody bound to the DNA polymerase. Inactivation of the Taq DNA Polymerase below 65°C prevents primer-dimer formation and non-specific amplification. This allows for specific amplification from low copy number target sequences.
What is the error rate of PCRBIO Taq DNA Polymerase?
The enzyme has an error rate of approximately 1 error per 2.0 x 10⁵ nucleotides incorporated.
What is the recommended extension time for PCRBIO Taq DNA Polymerase?
For amplicons between 1kb and 5kb, we recommend 15 seconds/kb for amplification from eukaryotic DNA. For shorter amplicons, a 1 second extension is sufficient.
More Information
PCRBIO Taq DNA Polymerase is a robust enzyme for all your everyday PCR applications including genotyping, screening and library construction. PCRBIO Taq DNA Polymerase performs consistently well on a broad range of templates including both GC and AT rich.
PCRBIO Taq DNA Polymerase has 5’-3’ exonuclease activities, but no 3’-5’ exonuclease (proofreading) activity. The enzyme has the same error rate as wild-type taq DNA polymerase, approximately 1 error per 2.0 x 105 nucleotides incorporated.
PCRBIO Taq DNA Polymerase provides the research community with an affordable routine application polymerase that performs to the highest possible standard, with a versatility that allows you to amplify with the highest speed, yield, specificity and consistency on the market. PCR products generated are A-tailed and may be cloned into TA cloning vectors. The enzyme is room temperature stable for 4 weeks and is produced using an enhanced 12 step purification strategy which includes physical, chemical and enzymatic removal of host DNA.
For added convenience, PCRBIO Taq DNA Polymerase is available as a ready-to-use 2x mix containing all reaction components except primers and template. PCRBIO Taq Mix Red contains a red dye suitable for direct loading and tracking during agarose gel electrophoresis.
Applications
- Routine application PCR
- TA cloning
- High throughput PCR
- Methylated DNA
- Crude sample PCR
- Standard and fast PCR
- Efficient specific amplification from GC and AT-rich sequences
Specifications
PCRBIO Taq DNA Polymerase
Component
500 Units
2000 Units
4000 Units
PCRBIO Taq DNA Polymerase (5u/μL)
1 x 100μL
4 x 100μL
8 x 100μL
5x PCRBIO Reaction Buffer
4 x 1mL
16 x 1mL
32 x 1mL
PCRBIO Taq Mix
Component
200 Reactions
1000 Reactions
2x PCRBIO Taq Mix
5 x 1mL
25 x 1mL
PCRBIO Taq Mix Red
Component
200 Reactions
1000 Reactions
2x PCRBIO Taq Mix Red
5 x 1mL
25 x 1mL
PCRBIO Taq DNA Polymerase
Component
PCRBIO Taq DNA Polymerase (5u/μL)
5x PCRBIO Reaction Buffer
500 Units
1 x 100μL
4 x 1mL
2000 Units
4 x 100μL
16 x 1mL
4000 Units
8 x 100μL
32 x 1mL
PCRBIO Taq Mix
Component
2x PCRBIO Taq Mix
200 Reactions
5 x 1mL
1000 Reactions
25 x 1mL
PCRBIO Taq Mix Red
Component
2x PCRBIO Taq Mix Red
200 Reactions
5 x 1mL
1000 Reactions
25 x 1mL
Reaction Volume
Storage
50μL
On arrival, products should be stored between -30 and -15°C. If stored correctly the kit will retain full activity for 12 months.
Reaction Volume
50μL
Storage
On arrival, products should be stored between -30 and -15°C. If stored correctly the kit will retain full activity for 12 months.
Documents
Product Manuals
Material Safety Data Sheets
Certificate of Analysis Finder
FAQs
Can I use PCRBIO Taq DNA Polymerase if my assay requires a specialized buffer?
The 5x PCRBIO Reaction Buffer supplied with PCRBIO Taq DNA Polymerase has been developed specifically for this enzyme and we highly recommend using them together. However, PCRBIO Taq DNA Polymerase should be compatible with any PCR buffer developed for use with wild-type Taq. If you use a customised buffer with PCRBIO Taq DNA Polymerase, keep in mind reaction parameters such as annealing temperature and concentrations of the enzyme, template, dNTPs and MgCl2, may require optimisation.
Can PCRBIO Taq DNA Polymerase be used for colony PCR?
Yes. If you’re working from bacterial colonies use a sterile tip to pick a colony and re-suspend into the 50µl PCR reaction. If working from liquid culture add 5µl of overnight culture to the final mix. Follow the general protocol and increase the initial denaturation time to 10 min at 95°C.
Can PCRBIO Taq DNA Polymerase be used to amplify DNA from blood samples?
We recommend using PCRBIO HS Taq DNA Polymerase for amplifying DNA from blood samples. If you’d like to use PCRBIO Taq DNA Polymerase, use 2 µL blood sample to a 50 µL PCR reaction and follow the general protocol. Please note that blood components may inhibit the PCR reaction. Perform a serial dilution of the sample in order to find the optimal template concentration for the PCR amplification.
Can PCRBIO Taq DNA Polymerase proofread?
No. PCRBIO Taq DNA Polymerase has 5’-3’ exonuclease activities, but no 3’-5’ exonuclease (proofreading) activity.
Does PCRBIO Taq DNA Polymerase generate an A-tail or blunt end on my PCR product?
PCR products generated with PCRBIO Taq DNA Polymerase are A-tailed and this makes it suitable for cloning into TA vectors. For further reading, refer to this literature1.
1 Yao, S., Hart, D. J. & An, Y. Recent advances in universal TA cloning methods for use in function studies. Protein Eng Des Sel, doi:10.1093/protein/gzw047 (2016).
My results contain a high background of non-specific amplicons or smears. What trouble-shooting suggestions do you have?
If smears are a concern, it’s good practice to ensure they are not an artifact of running agarose gel electrophoresis with sub optimal conditions. Sub optimal conditions can include high voltage or not allowing enough time for the gel to set1.
You may also need to troubleshoot the PCR reaction and consider the suggestions below2.
- Primers should be designed to prevent primer-primer interactions and improve specificity.
- Increase the annealing temperature or conducting an annealing temperature gradient PCR to determine the optimal annealing temperature.
- Reduce the amount of template in the reaction. For high quality DNA, use 1–100 ng genomic DNA or ≤5 ng plasmid/lambda DNA per 50 µL reaction.
- Reduce the number of cycles.
- Reduce the amount of enzyme per reaction.
- Reduce the primer concentration, but not lower than 100 nM of each primer.
- Include DMSO in the reaction to a final concentration of 5%–10%.
1 Koontz, L. Agarose Gel Electrophoresis. Laboratory methods in enzymology : DNA. First edition. edn, Vol. 529 35-45 (2013).
2 Lorenz, T. C. Polymerase chain reaction: basic protocol plus troubleshooting and optimization strategies. J Vis Exp, e3998, doi:10.3791/3998 (2012).
My results show a very low yield. What trouble-shooting suggestions do you have?
You may want to consider the suggestions below and also refer to the literature1.
- Optimise the annealing temperature in an annealing temperature gradient PCR.
- Increase the amount of template in the reaction.
- Increase the number of cycles.
- Increase the amount of enzyme per reaction.
- Increase the primer concentration, but do not exceed 1 µM of each primer.
- Try a fresh dNTP solution.
- Optimise the MgCl2
1 Lorenz, T. C. Polymerase chain reaction: basic protocol plus troubleshooting and optimization strategies. J Vis Exp, e3998, doi:10.3791/3998 (2012).
What is the apparent Mw of the Red Mix dye on agarose gels?
This non-inhibitory dye, added to enable direct gel loading, runs at a similar rate to 200-300 bp DNA fragments on a 1% agarose gel and at 50-100 bp on a 2% agarose gel. You may notice a shift in this apparent molecular weight when running gels of different agarose content.
What is the difference between PCRBIO Taq DNA Polymerase and PCRBIO HS Taq DNA Polymerase?
PCRBIO HS Taq DNA Polymerase uses proprietary antibody-mediated hot start technology whereas PCRBIO Taq DNA Polymerase does not have this feature. The interaction of the antibody with the Taq DNA Polymerase leaves the enzyme inactive until the hot start step.
The hot start refers to the initial activation step at 95°C, which subsequently deactivates the antibody bound to the DNA polymerase. Inactivation of the Taq DNA Polymerase below 65°C prevents primer-dimer formation and non-specific amplification. This allows for specific amplification from low copy number target sequences.
What is the error rate of PCRBIO Taq DNA Polymerase?
The enzyme has an error rate of approximately 1 error per 2.0 x 10⁵ nucleotides incorporated.
What is the recommended extension time for PCRBIO Taq DNA Polymerase?
For amplicons between 1kb and 5kb, we recommend 15 seconds/kb for amplification from eukaryotic DNA. For shorter amplicons, a 1 second extension is sufficient.